Recombination-aware phylogenetic analysis of Sarbecoviruses
The Sarbecoviruses are a group of single-stranded RNA viruses with a positive-sense genome, members of the Family Coronaviridae and the genus Betacoronavirus. SARS-CoV and SARS-CoV-2 are the most notable members of this group given their capacity to cause sever acute respiratory diseases in humans, with the latter also being the responsible for the ongoing pandemic that has taken the lives of more than 1 million people.
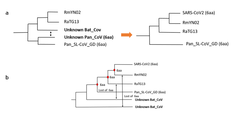
After the start of the SARS-CoV-2 pandemic in late 2019, several research groups from around the world started the task of clarifying the origin of this virus, with one particular region of the genome, the Spike or S gene, occupying the center of the discussion. The Spike protein has a central role in facilitating human infection, since it can recognize the ACE2 receptors of the human cells allowing the binding and introduction of the virus into the cell. Interestingly, a region located within the Receptor Binding Domain (RBD) of the S gene presents six amino acid substitutions that are common to only SARS-CoV-2 and a pangolin coronavirus sampled in China in 2019, while full genome phylogenetic analysis have shown that SARS-CoV-2 is more similar to viruses from bats. This suggests that SARS-CoV-2 could have originated after a recombination event between bat and pangolin coronaviruses; while an alternative hypothesis, states that these substitutions were already present in the most recent common ancestor of these two and the next closest bat coronavirus (which lacks five of the six substitutions).
In this project, we test the suggested scenarios regarding the origin of the RBD region in SARS-CoV-2 and its closest relatives. This will be done using the BEAST2 package Bacter, which allows the joint estimation of the phylogeny and the recombination history, including ancestral recombination events.











